Repopulating a table after bug fix
Let's work out an example of how to proceed whenever we need to change some rule, resulting in a need to repopulate one of the tables in the ToMCAT dataset.
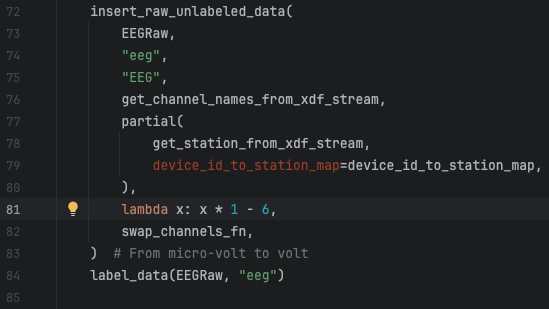
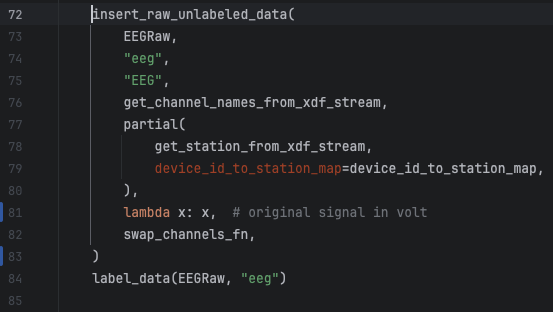
This is a real example of a change I needed to do in the raw EEG signals that were being converted to microvolt before persisted in the table eeg_raw. So, to fix this, I need to remove that conversion from the code and repopulate the eeg_raw and eeg_sync tables to reflect the unconverted values. The eeg_sync table entries need to be repopulated too because its content is derived from the entries in the eeg_raw table.
Note: all code changes steps below are in a new development branch. Scripts, however, were executed in prod mode, meaning they affected the Postgres database directly.
Step 1: Fix the bug (or change the logic)
This is an easy fix in line 81 of the file human_experiments/datasette_interface/datasette_interface/raw/process_eeg_raw_data.py.
Step 2: Pull the new code in Gauss
On Gauss, git pull and switch to the development branch we pushed the code change in the previous step to. Also, make sure to activate your virtual environment in Gauss so we can execute the script to repopulate the EEG table.
Step 3: Repopulate the tables
Before we run the script to update the eeg tables, we need to erase their content. This step is necessary because the table population script skips group sessions for which we already have data in the table. It was developed to be called incrementally, every time we run new experiments and have new data available.
Delete the content of the eeg tables
You can do this in a client application running locally on your machine and connected to the database server on Gauss or by connecting to the database directly in Gauss. The instructions below shows the latter.
# Run the following commands in a terminal connected to Gauss.
# The first command will connect to the database and open an interface from which we can call SQL commands.
# ekg and gsr sync tables need to be cleaned as well because raw EKG and GSR signals are columns in the eeg_raw table.
# Instead of deleting the contents, I prefer to drop the table which is quicker.
psql -p 5433 -h /space/paulosoares/postgres/run -d tomcat
drop table eeg_raw;
drop table eeg_sync;
drop table ekg_sync;
drop table gsr_sync;Repopulate eeg_raw from scratch
Now that the table is clean, we can use one of our make commands to repopulate the table with the commands below.
# Be sure you are under the directory human_experiments/datasette_interface
# Replace db_pass with your Postgres password
# We need to first recreate the eeg tables because we dropped them in the last step.
# The command below will only create tables and indices that do not exist in the database.
# It won't affect existing tables.
working_env=production db_pass=<user_postgres_pass> make create_tables
# We use the TBS environment variable to reduce the data entitites we want to process. In this case, only eeg was affected.
# For a full list of entities available, do PYTHONPATH="." ./bin/populate_raw_tables.py -h
working_env=production db_pass=<user_postgres_pass> TBS="eeg" make update_rawRepopulate sync tables from scratch
This step can only be executed after the former because the script to generate synchronized signals reads from the raw signals table.
# Be sure you are under the directory human_experiments/datasette_interface
# Replace db_pass with your Postgres password
# We use the N_JOBS environment variable to define the number of group sessions we want to process in parallel
working_env=production db_pass=<user_postgres_pass> N_JOBS=40 make sync_eeg
working_env=production db_pass=<user_postgres_pass> N_JOBS=40 make sync_gsr
working_env=production db_pass=<user_postgres_pass> N_JOBS=40 make sync_ekgStep 4: Copy contents to the sqlite database
After the Postgres dataset is updated, we need to copy the new content to our sqlite database which we use to publish our dataset online. I currently have a copy of this dataset under /space/paulosoares/tomcat/tomcat.db which is the one I will update with the commands below. Later, this is copied to a final location where our dataset’s webpage links to.
Note: this is going to take a long time, even days. So it’s paramount we run this in a TMUX session if we haven’t been doing so with the previous commands. There’s a progress indicator but, after reaching 100%, you still may have to wait for a long time because of an operation called VACUUM we cannot disable with the postgres to sqlite library we are using. This operation optimizes the sqlite db file by removing empty segments. Those empty segments are mostly caused by deletions in the database which we do not have since our dataset is mostly readonly. So, there’s little to be optimized in space in our case thus it would be ideal if we could find a way to prevent this operation in the future.
# RUN IN A TMUX SESSION!!!
# The command below reads from the Postgres table by default so we don't need to set the working_env variable.
# Enlist the tables we want to copy with the TBS working variable.
TBS="eeg_raw, eeg_sync, ekg_sync, gsr_sync" make to_sqlite